Ligand Binding Sites & Predictions
Data for nonpolymer-ligand binding sites across deposited structures and predictions derived from it.
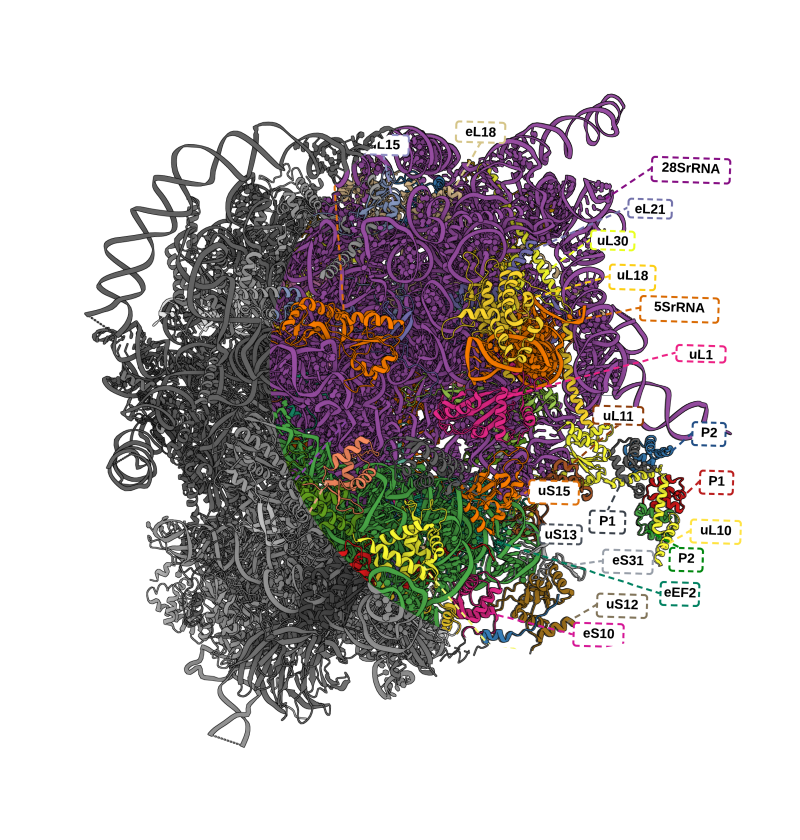
Multiple classes of antibiotics and their binding sites identified on the structure 7K00 (due to Watson et al., 2021) of the E. coli ribosome.





